
The Interpretation Space is a companion to the Annotation space that allows clinicians and researchers to take detailed notes about annotation data at the variant, sample, and gene level for the purpose of genomic variant classification.
Context
The interpretation space is a tool that helps clinicians and researchers keep a record of which genetic variants from a particular case are most likely to be pathogenic or benign. Here, clinicians and researchers are able to write notes, view and invoke ACMG rules according to the standard guidelines or their own custom ones, can generate an auto-classification, and record their final variant classifications. Paired with the CGAP annotation space, the interpretation space allows clinicians to use the annotation data for samples, genes, and variants to build, record, and justify their variant classifications for future reference.
My Role
Collaborators
Organization
Technology Used
JS(ES6), React, React-Bootstrap, SASS/SCSS
Timeframe
April - May 2021
Design Phase
The design of this page and all of the features on it was primarily created by Shannon Ehmsen. However, I was involved in both the design and development stages of CGAP's interpretation space. Our graphic designer created wireframes and mock-ups, in communication with us front-end developers and the clinicians on our user test team. We provided technical feasibility feedback, feature ideas, and general UI/UX feedback along the way. The clinicians and researchers we work with also provided subject matter expertise that helped us to empathize with their needs, and address the peculiarities of both distinct use-cases.
Implementation Process
Once the mockups were complete, they were handed off to the data wranglers, who in communication with us, made the necessary schema updates to support the changes. Once those changes were in, they passed their branch off to me, and I built the interface with React and SCSS.
In order to develop the algorithm for auto-classifying variants, I read numerous papers regarding the American College of Medical Genetics (ACMG) Guidelines for the classification of genomic variants (especially this one) and worked closely with clinicians on the Brigham Genomic Medicine team to ensure the results were accurate. For context: the ACMG Guidelines are a standardized set of rules for when to classify a genetic variant as being benign or pathogenic. If a genetic variant, for instance, has a high allele frequency (so, it's found often in the general population), that is a piece of evidence that it might be benign. Enough evidence that supports benign or pathogenic classification, and that classification becomes better supported for use in clinical or research contexts.
However, some labs (especially those doing novel research) may have their own adjustments they make to certain rules. So, they may want to weigh certain rules more or less. It was extremely important that users could make these adjustments on the fly, if they so desired. We do want to, in the future, make it possible to set defaults on a project by project basis, for this use-case, but that wasn't in the scope of this version.
Anyway, I developed several iterations of the note-taking interface, as we discovered new user needs and sticking points, some of which are discussed further in the Retrospective and Takeaways section below.
Features
Since Interpretation Space is primarily a note-taking interface, most of the features I implemented are pertaining to that:
Tabbed menu for swapping between different types of notes
Note saving and unsaved state indicators (+warning when navigating away)
ACMG rule invocation for clinical users with ability to modify rule strength
Auto-classification according to the ACMG guidelines
Custom classifications and means of saving those
Hiding and showing interpretation space depending on how user starts analysis
And all of this is not even to mention the data-rich UIs we developed for the Annotation Space itself, including the Gene, Variant, and Sample tabs (all of which I developed together with Alex Balashov), which provide data for use in the interpretation process in a compact and easily scannable interface.
Retrospective and Takeaways
The biggest setback we faced on this project was actually a really tiny UX detail that we overlooked in the planning phases. Originally, we planned for the same button (with dropdown) to be used to handle various ways of saving (to the case or the knowledge base, for instance, which basically just allow different levels of access to the note):
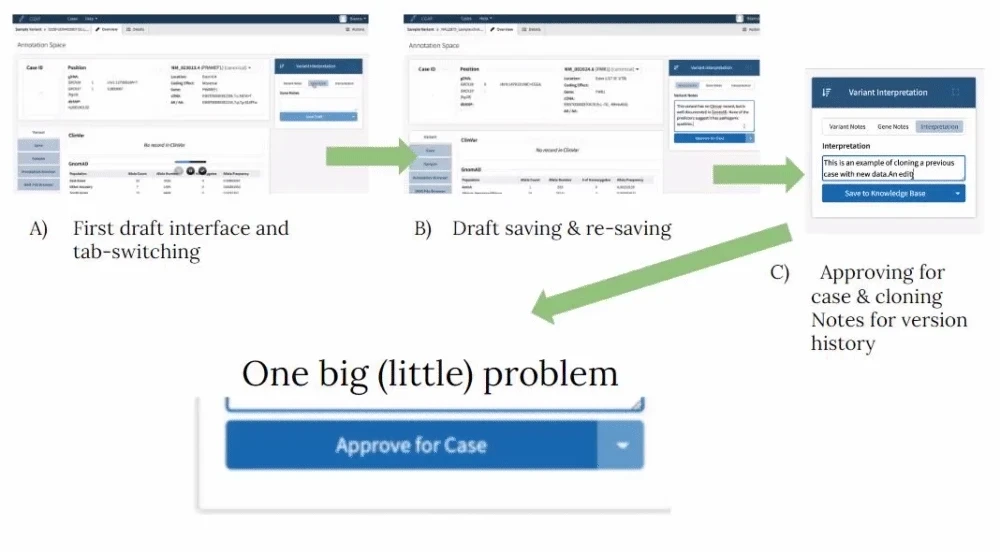
In a clinical workflow, the primary concern is always accuracy; our testers were very worried about the possibility of people saving a draft, having the "approve for case" version of the button activate, and then accidentally approving the note before it was ready. This concern was even greater when "approving for case", since updating the same button meant users could slip and add something to the global knowledge base. At this point in the process, there was no easy way to revert either of these sorts of changes, and we didn't have the time to add those "undo" features before release, so instead adjusted the plan quite a bit.
In the end, we decided on a two-button workflow: one button for saving drafts, and one for moving back to the case. And then the entirety of the note approving and sharing functionality was moved to a dedicated interface, where it would be harder to make these sorts of mistakes, and easier to remedy them should they happen.
This is a really great example of the value of including interactive prototypes in the design phase after the creation of static mockups, but before the coding phase. Small interaction details like this can be easily missed when you don't actually get to "feel" what it is like to click between different parts of an interface.
Outcome
In the end, it took us a little longer than initially expected to put together this page and all of its features, but it is one of the most frequently used features on the CGAP platform (aside from search and filter blocks), for both single-nucleotide and structural variant interpretation and across research and clinical users.
Demo
Dr. Joel Krier, one of the clinicians that has helped us shape the product's experience and marketing, did a walk-through of the interpretation and annotation spaces showing how the tools would be used in a typical case analysis:
You can also try out CGAP for yourself by creating an account on our training instance. To do that, go to the link below and attempt to log in with the credentials you'd like to use with the platform; you'll be prompted to register.



